Renjie Hu
Benchmarking and Enhancing PPG-Based Cuffless Blood Pressure Estimation Methods
Feb 04, 2026Abstract:Cuffless blood pressure screening based on easily acquired photoplethysmography (PPG) signals offers a practical pathway toward scalable cardiovascular health assessment. Despite rapid progress, existing PPG-based blood pressure estimation models have not consistently achieved the established clinical numerical limits such as AAMI/ISO 81060-2, and prior evaluations often lack the rigorous experimental controls necessary for valid clinical assessment. Moreover, the publicly available datasets commonly used are heterogeneous and lack physiologically controlled conditions for fair benchmarking. To enable fair benchmarking under physiologically controlled conditions, we created a standardized benchmarking subset NBPDB comprising 101,453 high-quality PPG segments from 1,103 healthy adults, derived from MIMIC-III and VitalDB. Using this dataset, we systematically benchmarked several state-of-the-art PPG-based models. The results showed that none of the evaluated models met the AAMI/ISO 81060-2 accuracy requirements (mean error $<$ 5 mmHg and standard deviation $<$ 8 mmHg). To improve model accuracy, we modified these models and added patient demographic data such as age, sex, and body mass index as additional inputs. Our modifications consistently improved performance across all models. In particular, the MInception model reduced error by 23\% after adding the demographic data and yielded mean absolute errors of 4.75 mmHg (SBP) and 2.90 mmHg (DBP), achieves accuracy comparable to the numerical limits defined by AAMI/ISO accuracy standards. Our results show that existing PPG-based BP estimation models lack clinical practicality under standardized conditions, while incorporating demographic information markedly improves their accuracy and physiological validity.
A Reproducible Framework for Bias-Resistant Machine Learning on Small-Sample Neuroimaging Data
Feb 02, 2026Abstract:We introduce a reproducible, bias-resistant machine learning framework that integrates domain-informed feature engineering, nested cross-validation, and calibrated decision-threshold optimization for small-sample neuroimaging data. Conventional cross-validation frameworks that reuse the same folds for both model selection and performance estimation yield optimistically biased results, limiting reproducibility and generalization. Demonstrated on a high-dimensional structural MRI dataset of deep brain stimulation cognitive outcomes, the framework achieved a nested-CV balanced accuracy of 0.660\,$\pm$\,0.068 using a compact, interpretable subset selected via importance-guided ranking. By combining interpretability and unbiased evaluation, this work provides a generalizable computational blueprint for reliable machine learning in data-limited biomedical domains.
NeuroMoE: A Transformer-Based Mixture-of-Experts Framework for Multi-Modal Neurological Disorder Classification
Jun 17, 2025Abstract:The integration of multi-modal Magnetic Resonance Imaging (MRI) and clinical data holds great promise for enhancing the diagnosis of neurological disorders (NDs) in real-world clinical settings. Deep Learning (DL) has recently emerged as a powerful tool for extracting meaningful patterns from medical data to aid in diagnosis. However, existing DL approaches struggle to effectively leverage multi-modal MRI and clinical data, leading to suboptimal performance. To address this challenge, we utilize a unique, proprietary multi-modal clinical dataset curated for ND research. Based on this dataset, we propose a novel transformer-based Mixture-of-Experts (MoE) framework for ND classification, leveraging multiple MRI modalities-anatomical (aMRI), Diffusion Tensor Imaging (DTI), and functional (fMRI)-alongside clinical assessments. Our framework employs transformer encoders to capture spatial relationships within volumetric MRI data while utilizing modality-specific experts for targeted feature extraction. A gating mechanism with adaptive fusion dynamically integrates expert outputs, ensuring optimal predictive performance. Comprehensive experiments and comparisons with multiple baselines demonstrate that our multi-modal approach significantly enhances diagnostic accuracy, particularly in distinguishing overlapping disease states. Our framework achieves a validation accuracy of 82.47\%, outperforming baseline methods by over 10\%, highlighting its potential to improve ND diagnosis by applying multi-modal learning to real-world clinical data.
Spiking Networks for Improved Cognitive Abilities of Edge Computing Devices
Dec 19, 2019
Abstract:This concept paper highlights a recently opened opportunity for large scale analytical algorithms to be trained directly on edge devices. Such approach is a response to the arising need of processing data generated by natural person (a human being), also known as personal data. Spiking Neural networks are the core method behind it: suitable for a low latency energy-constrained hardware, enabling local training or re-training, while not taking advantage of scalability available in the Cloud.
Macro action selection with deep reinforcement learning in StarCraft
Dec 02, 2018

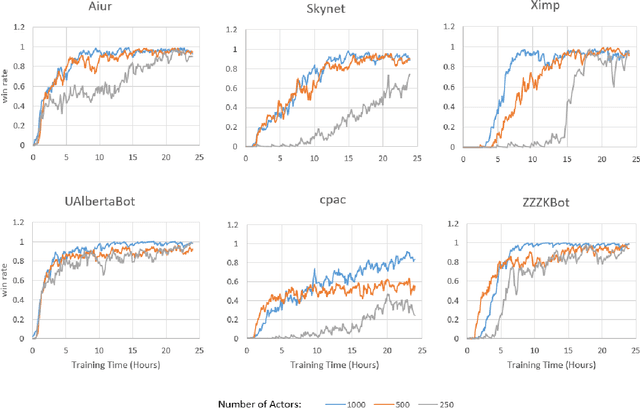
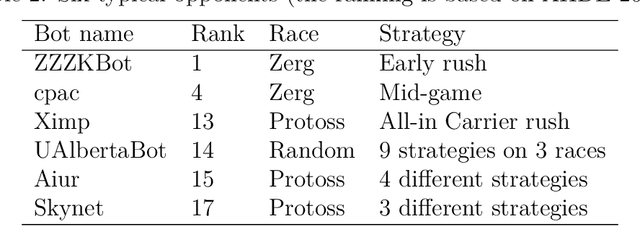
Abstract:StarCraft (SC) is one of the most popular and successful Real Time Strategy (RTS) games. In recent years, SC is also considered as a testbed for AI research, due to its enormous state space, hidden information, multi-agent collaboration and so on. Thanks to the annual AIIDE and CIG competitions, a growing number of bots are proposed and being continuously improved. However, a big gap still remains between the top bot and the professional human players. One vital reason is that current bots mainly rely on predefined rules to perform macro actions. These rules are not scalable and efficient enough to cope with the large but partially observed macro state space in SC. In this paper, we propose a DRL based framework to do macro action selection. Our framework combines the reinforcement learning approach Ape-X DQN with Long-Short-Term-Memory (LSTM) to improve the macro action selection in bot. We evaluate our bot, named as LastOrder, on the AIIDE 2017 StarCraft AI competition bots set. Our bot achieves overall 83% win-rate, outperforming 26 bots in total 28 entrants.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge